
Artificial intelligence and machine learning can help users sift through terabytes of data to arrive at a conclusion driven by relevant information, prior results, and statistics. But how much should we trust those conclusions—especially given AI’s tendency to hallucinate, or simply make things up?
Consider the use of an AI health care system that predicts a patient’s medical diagnosis or risk of disease. A doctor’s choice to trust that AI’s advice can have a major impact on the patient’s health outcomes; in such high-stakes scenarios, the cost of a hallucination could be someone’s life.
AI predictions can vary in trustworthiness, for any number of reasons: maybe the data are bad to begin with, or the system hasn’t seen enough examples of the current situation it’s facing to have sufficiently learned what to do in that scenario. Additionally, most of the modern deep neural networks behind AI predict singular outcomes—that is, whichever has the “highest score” based on a particular model. In the context of health care, this would mean the aforementioned AI system would return only one probable disease diagnosis out of the many possibilities. But just how certain is the AI that it’s got the correct answer?
Uncertainty quantification methods attempt to discover just that, generally by prompting ML models to return a set of possible outcomes—the larger the predicted set, the more uncertain that model would be in its prediction. Now, Hopkins computer scientists are making breakthroughs in refining such UQ methods for use in medical imaging and other real-world scenarios.
Trusting ML Medical Imaging Reconstructions
Diffusion models are capable of generating varied and high-quality images and are employed by apps like DALL-E and Midjourney. Aside from generating artwork, they can also be used in medical imaging to denoise and reconstruct a patient’s CT or MRI scan, allowing for lower radiation doses during the scan itself.
“Although these systems do produce high-quality reconstructions, we currently have no tools to guarantee the model did not hallucinate some of the more fine-grained details,” says Jacopo Teneggi, a doctoral candidate in the Department of Computer Science and an affiliate of the Johns Hopkins Mathematical Institute for Data Science. “In other words, how can we be sure the reconstructed images are indeed the true anatomy of the patient?”
Under the guidance of his advisor Jeremias Sulam, an assistant professor of biomedical engineering and faculty at MINDS, Teneggi sought to answer how certain a diffusion model is in its reconstructions and to determine how far those reconstructions are from a patient’s true anatomy. His effort was joined by I-STAR Lab members J. Webster Stayman, an associate professor of biomedical engineering and electrical and computer engineering, and Matthew Tivnan, Engr ‘23 (PhD), now an investigator at Harvard Medical School and Massachusetts General Hospital.
The multidisciplinary team used the ideas of conformal prediction and conformal risk control to quantify the uncertainty of a diffusion model with valid statistical guarantees. They demonstrated their novel high-dimensional UQ algorithm’s state-of-the-art performance on denoising real-world abdominal CT scans.
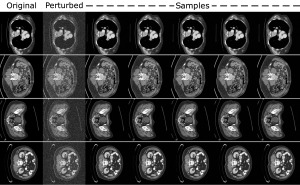
Example abdominal scan images sampled via the team’s algorithm.
“Practically, we can now provide radiologists with reconstructions and uncertainty maps that precisely guarantee that the true anatomy of a future patient will be contained within the model’s uncertainty intervals—without knowing what that true anatomy is,” Teneggi explains.
Future work for the team entails extending their uncertainty intervals to semantic features, rather than individual pixels, and improving optimization schemes.
“We envision our tools contributing to the responsible use of ML in modern settings,” says Teneggi.
Their paper was accepted to the 2023 International Conference on Machine Learning in Honolulu, Hawaii, and the team will be presenting their work as a poster at the Radiological Society of North America’s Annual Meeting taking place November 26–30 in Chicago, Illinois.
Flexible UQ Methods for the Real World
Two key challenges still limit the real-world use of UQ in AI and ML. The first is caused by discrepancies between the data a model is trained on versus the data it sees at deployment. The second is that many UQ approaches require either intensive computing power or an impractical amount or quality of data that may simply be unavailable in real-world scenarios.
But a group of Hopkins researchers—Suchi Saria, the John C. Malone Associate Professor of Computer Science and founding research director of the Malone Center for Engineering in Healthcare; Anqi “Angie” Liu, an assistant professor of computer science; and Drew Prinster, a PhD candidate in the Department of Computer Science—recently presented a novel collection of UQ methods for complex AI and ML models. Called “JAWS-X,” their proposed methods promise to flexibly balance computational and data-use efficiency for practical UQ deployment.
“This is especially important when data are scarce or costly to collect, such as in health care, where patient privacy considerations rightfully limit data use, or in biomolecular design, where data collection requires expensive and time-consuming experimental procedures,” Prinster explains.
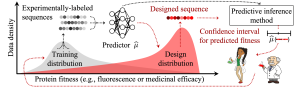
Illustration of a biomolecular design data shift scenario.
Building on prior work published at the 2022 Conference on Neural Information Processing Systems, the team says they have generalized previous UQ methods to allow for real-world data shifts while also striking a flexible, favorable balance between computing requirements and data-use efficiency.
A technique called “importance weighting” allows the JAWS-X UQ methods to adapt their uncertainty estimates in response to data shifts, the team explains. In the example of an AI health care system encountering different patient demographics than what it was trained on, the JAWS-X methods account for this shift by emphasizing the information from the minority-group patients they have seen before in the training data and de-emphasizing information from the majority group in the training data.
And where previous methods offered a “pick-your-poison” choice between either prohibitive computational costs or inefficient—and potentially harmful—data use, the team claims their JAWS-X methods avoid this false dichotomy by allowing users to flexibly adapt resource requirements needed for their specific UQ problem. In other words, depending on the computing power available to researchers, they can choose a JAWS-X method to match so that they achieve a balance between model performance and fast computation.
The Hopkins computer scientists report experimental results demonstrating that their JAWS-X methods lead to substantial improvements as compared to state-of-the-art work in the efficiency and informativeness of AI and ML UQ in a fluorescent protein design use case.
“This is an important step toward a framework for building appropriate trust dynamics in human-AI teams,” says Prinster.
Their paper was not only accepted to ICML 2023, but was additionally selected for an oral presentation at the prestigious conference; this designation is awarded to only the top 2 percent of conference submissions.