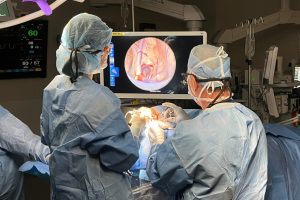
Johns Hopkins researchers have improved their cutting-edge augmented endoscopy algorithm—which allows neurosurgeons to navigate the deep brain in real time without having to use invasive clamps or pins—for real-life operations by integrating AI tracking. Their new method, called R2D2-E, can follow specific anatomical features in an endoscopic video even in the presence of blurring, glare, and other visual flaws, promising better neurosurgical accuracy and patient outcomes.
The researchers’ work appears in IEEE Transactions on Biomedical Engineering.
Previously, the team tested its original navigation algorithm on a “phantom,” or a realistic brain dummy. But in real patients, blood and other fluids often obscure an endoscope’s view, and the lighting required to see deep inside the brain can cause glare, reflections, and other visual artifacts that challenged the algorithm’s ability to discern different anatomical features.
Deep learning can help account for these flaws, but training these methods traditionally requires more ground-truth data—specifically, hand-labeled annotations noting the correlation of specific points between various surgical video frames—than is typically available in neuroendoscopies.
“Rather than depending on this kind of time-consuming annotation, we simulated our own physical and visual artifacts on images from 15 real-life neuroendoscopies to develop a new AI-based method, R2D2-E,” says first author Prasad Vagdargi, Engr ’25 (PhD). “Then we trained a neural network on smaller, cropped portions of these image pairs—the original and the simulated—to be able to match them.”
The researchers report that R2D2-E can match the features of even the tiniest of blood vessels and anatomical edges for both simple and complex images. Their method also generates confidence scores for each identified feature based on how repeatable and reliable the algorithm thinks the feature is. Only anatomical features with the highest confidence scores are used for tracking between video frames, ensuring consistent localization and reconstruction.
The team evaluated R2D2-E against state-of-the-art methods and even used it to create a 3D point-cloud from a real neuroendoscopic video, measuring how accurate the reconstruction was compared to a patient’s original MRI scan.
“Our findings suggest that R2D2-E not only outperformed the other evaluated methods in terms of accuracy by up to 25%, but also provided a more practical approach to feature detection and matching in real-life applications,” Vagdargi says.
Such applications include creating a neuroendoscopic overlay that displays target anatomy and critical arteries to avoid based on a patient’s MRI, as pictured below, to help neurosurgeons navigate the deep brain during an operation—even beyond their current field of view.
“R2D2-E enables accurate, real-time 3D reconstruction in neuroendoscopy, offering robust feature detection in the presence of endoscopic artifacts and providing up-to-date navigation following soft-tissue deformation,” Vagdargi says. “Our method advances the capabilities of vision-based guidance and augmented visualization of target structures in neuroendoscopic procedures, supporting future integration with real-world clinical systems.”
Senior and corresponding author of this work is Jeffrey H. Siewerdsen, a professor of biomedical engineering and faculty in and director of the Institute for Data Science in Oncology at the University of Texas MD Anderson Cancer Center. Coauthors include Mandell Bellmore Professor of Computer Science Gregory D. Hager; Craig Jones, an assistant research professor in the Department of Computer Science; and researchers from the Whiting School’s Department of Biomedical Engineering, the Departments of Radiation Oncology and Neurology and Neurosurgery at the Johns Hopkins Hospital, and Medtronic.
This work was supported by NIH U01 Grant NS107133.