The significance of endoscopic visualization and navigation is the introduction of a paradigm shift in surgical navigation by using a device present in every endoscopic surgery - the endoscope - to improve registration and visualization of anatomy. This has numerous positive impacts. Most importantly, it provides an inexpensive, noninvasive and radiation-free method to enhance registration accuracy at any point during the procedure. Enhancements in registration reduce ambiguity for the surgeon during surgery, increase confidence, and improve workflow by reducing the need to reregister or reimage the patient. This accrues additional benefits to the patient and hospital by reducing the level of radiation exposure and cost by eliminating the need for intraoperative CT imaging. The endoscope can also be used as a measurement device to update anatomic models during a procedure, improving the ability of the surgeon to visualize the progress of the surgery.
The main areas of research being explored in order to achieve these objectives are detailed here:
Segmentation and modeling of anatomical structures from CT scans
-
Patient head CT scans acquired preoperatively can be preprocessed in order to segment relevant structures and regions of interest for preoperative planning as well as intraoperative navigation. There are severl methods that can be used to compute these segmentations. We use a method that gives us correspondence between segmentations in different CT scans for free.
We use a template CT image to make manual labels which can be extracted as 3D triangular meshes. These meshes are then transfered to any new CT scan using deformation fields obtained from diffeomorphic deformable registration between the template and the new CT scan. This can be repeated for any number of CT scans, producing a set of segmented 3D meshes with correspondences. These correspondences can be exploited to build statistical shape models (SSMs) of shapes using a method like principle component analysis (SNH16).
However, this assumes that segmentations obtained from deformable registration are perfect, which is often not true. Segmentations can be improved by using gradients in the CT image to pull vertices in the mesh towards edges. Statistics can be obtained using these updated shapes. However, this assumes that independently moving the vertices of each shape in the dataset does not cause deterioration in correspondences, which is also not true. Therefore, the segmentation improvement can be modified by constraining the movement of vertices using SSMs (LEO18).
Surface shape estimation from endoscopic video
-
Feature extraction from endoscopic video frames is a challenging problem. Several methods have attempted to extract features from regular world images and videos. These images and videos are generally highly textured and contain several sharp features like straight lines and corners. However, human anatomy, such as the nasal cavity, is not feature-rich, making it difficult to find and match features from different video frames.
Methods that work in regular world scenarios have been applied to endoscopic videos. Careful implementation of such methods can yield sparse 3D point clouds, which only enables a rough estimation of the underlying surface shape (LEO16). However, these sparse point clouds registered to CT models can be used to learn the depth at the corresponding pixels in the 2D images under some explicit reflectance model. Based on the learned model, a depth value can be assigned to each pixel in the video frames (REI16).
This work has more recently been extended to estimate dense depth directly from sparse point clouds without registration to CT models. This self-supervised deep learning method ensures depth consistency between frames using relative camera motion in order to avoid learning erroneous depth estimates from outliers in the sparse point cloud (LIU18).
Video-CT registration
-
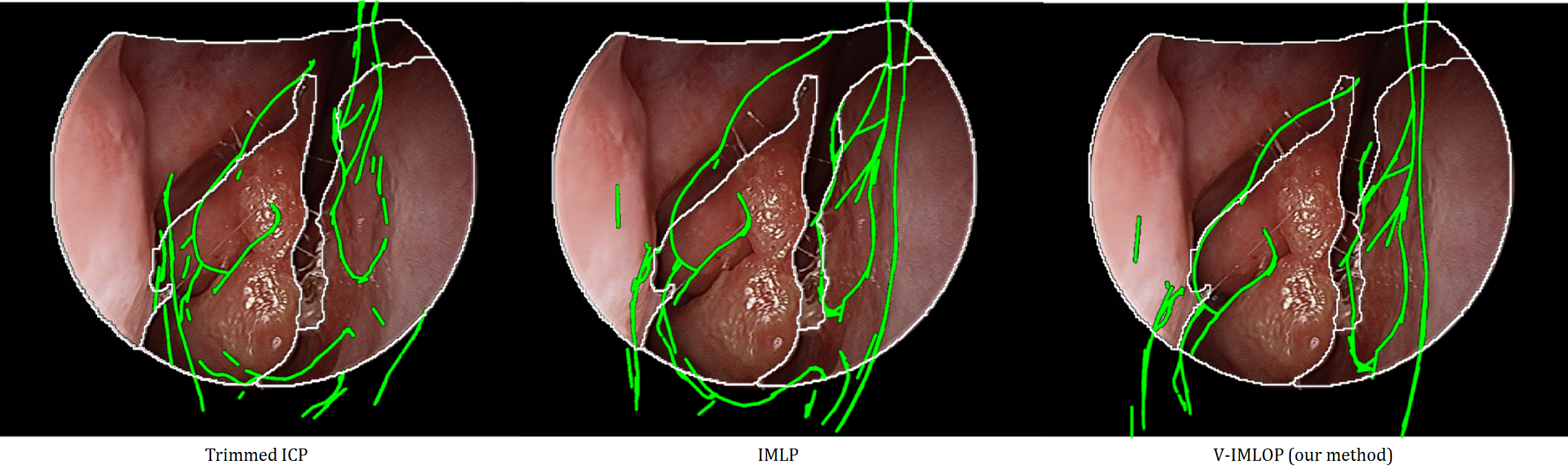
Features extracted from CT scans and endoscopic video frames can be matched and aligned using registration techniques in order to enhance endoscopic videos with additional information from the CT scans. This is crucial since endoscopic videos have limited field of view, and using registered information from CT scans can inform clinicians about nearby critical structures that may be occluded or not in the view of the endoscope.
Several different approaches can be taken to compute the registration between CT and video features. One approach is to use the iterative closest point (ICP) algorithm and solve for a rigid transformation (rotation and translation) between point features from video and CT. However, the scale of the features from video is often unknown, and, therefore, solving for an affine tranformation (rotation, translation and scale) is more practical. Using sparse point clouds extracted from video, submillimeter registration can often be achieved between video and CT (LEO16). However, a registration algorithm can often show submillimeter registation error when the true registration error is higher. This happens when the endoscope is in a narrow tube like structure with few identifying features. Therefore, understanding the stability of the computed registration can tell the clinicans something about how reliable a computed registration may be (SNH18). In order to better inform the algorithm and compute more reliable registrations, additional features can be used. Using oriented occluding contours in addition to point features helps remove ambiguities in registrations, further improving registration outcomes (SNH17).
Often, CT images are not avaiable in clinic or are too low resolution to be used for surgery. In order to still be able to provide contexual information aligned with video, SSMs can be deformably registered to video features to produce an approximate shape of patient anatomy which is registered to video (LIU18).
Confidence assignment for registration outcomes
-
More information on this coming soon!
Related publications
[LIU18] X Liu, A Sinha, M Unberath, M Ishii, GD Hager, RH Taylor, A Reiter. “Self-supervised Learning for Dense Depth Estimation in Monocular Endoscopy”, Lecture Notes in Computer Science, Vol. 11041, pp 128-138, Computer Assisted and Robotic Endoscopy — CARE 2018 (MICCAI Workshop), Granada, Spain (October 02, 2018)
[SNH18] A Sinha, X Liu, A Reiter, M Ishii, GD Hager, RH Taylor. “Endoscopic navigation in the absence of CT imaging”, Lecture Notes in Computer Science, Vol. 11073, pp 64-71, Medical Image Computing and Computer-Assisted Intervention — MICCAI 2018, Granada, Spain (September 13, 2018)
[LEO18] S Leonard, A Sinha, A Reiter, M Ishii, GL Galia, RH Taylor, GD Hager. “Evaluation and Stability Analysis of Video-Based Navigation System for Functional Endoscopic Sinus Surgery on In-Vivo Clinical Data”, IEEE TMI, Early Access (May, 2018)
[SNH17] A Sinha, A Reiter, S Leonard, M Ishii, RH Taylor, GD Hager. “Simultaneous segmentation and correspondence improvement using statistical modes”, Proc. SPIE, 10133, Medical Imaging 2017: Image Processing, 101331B, Orlando, FL (February 24, 2017)
[BIL16] SD Billings, A Sinha, A Reiter, S Leonard, M Ishii, GD Hager, RH Taylor. “Anatomically Constrained Video-CT Registration via the V-IMLOP Algorithm”, Lecture Notes in Computer Science, Vol. 9902, pp 133-141, Medical Image Computing and Computer-Assisted Intervention — MICCAI 2016, Athens, Greece (October 02, 2016)
[REI16] A Reiter, S Leonard, A Sinha, M Ishii, RH Taylor, GD Hager. “Endoscopic-CT: learning-based photometric reconstruction for endoscopic sinus surgery”, Proc. SPIE 9784, Medical Imaging 2016: Image Processing, 978418, San Diego, CA (March 21, 2016)
[LEO16] S Leonard, A Reiter, A Sinha, M Ishii, RH Taylor, GD Hager. “Image-based navigation for functional endoscopic sinus surgery using structure from motion”, Proc. SPIE 9784, Medical Imaging 2016: Image Processing, 97840V, San Diego, CA (March 21, 2016)
[SNH16] A Sinha, S Leonard, A Reiter, M Ishii, RH Taylor, GD Hager. “Automatic segmentation and statistical shape modeling of the paranasal sinuses to estimate natural variations”, Proc. SPIE 9784, Medical Imaging 2016: Image Processing, 97840D, San Diego, CA (March 21, 2016)
Funding sources
National Institutes of Health Grant No. R01 EB015530: Enhanced Navigation for Endoscopic Sinus Surgery through Video Analysis, 7/1/2012-6/30/2017





