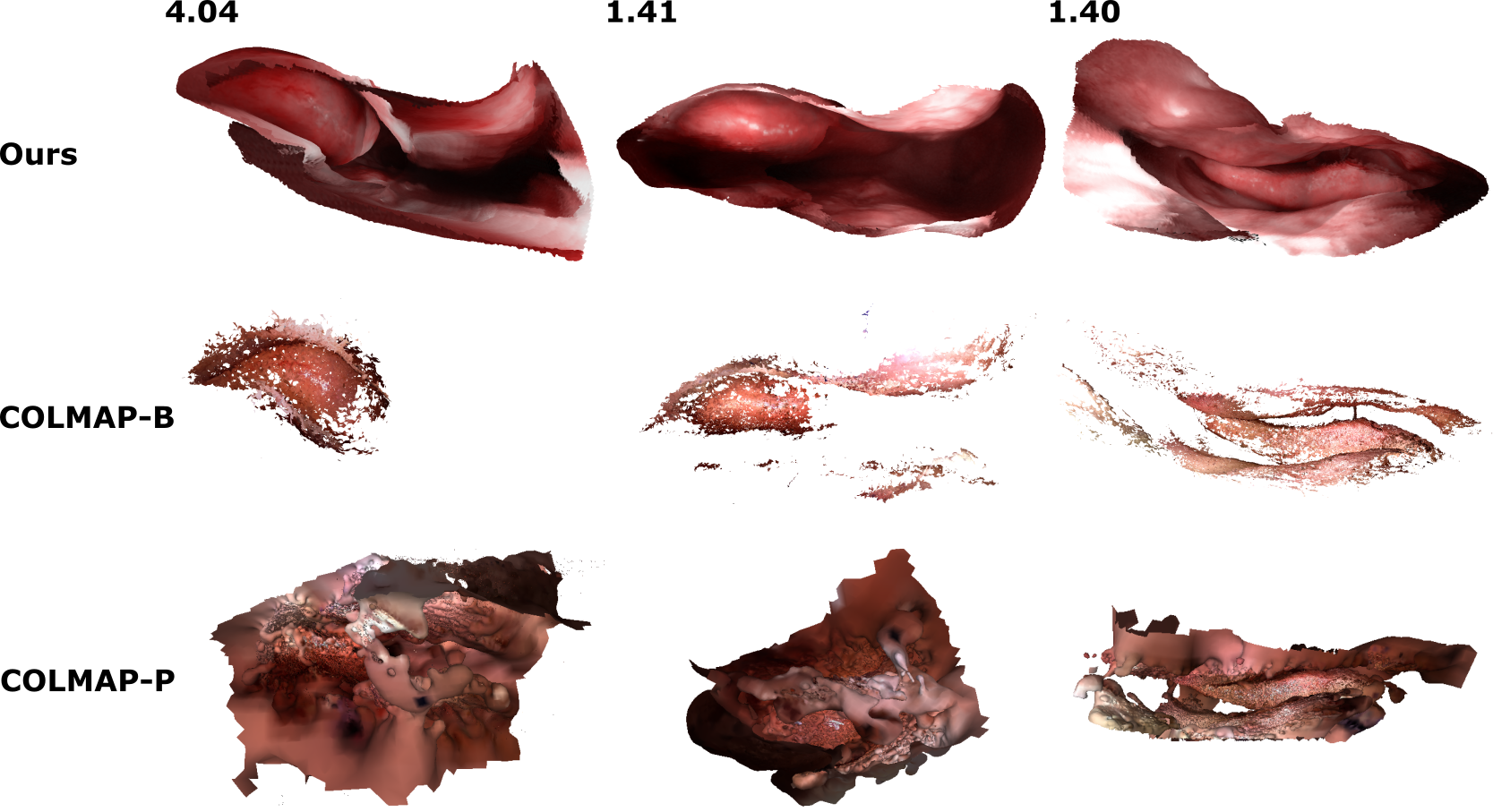
Reconstructing Sinus Anatomy from Endoscopic Video
We present a patient-specific, learning-based method for 3D
reconstruction of sinus surface anatomy directly and only from
endoscopic videos. We demonstrate the effectiveness and
accuracy of our method on in and ex vivo data where we compare
to sparse reconstructions from Structure from Motion, dense
reconstruction from COLMAP, and ground truth anatomy from CT.
Our textured reconstructions are watertight and enable
measurement of clinically relevant parameters in good
agreement with CT. (MICCAI 2020)
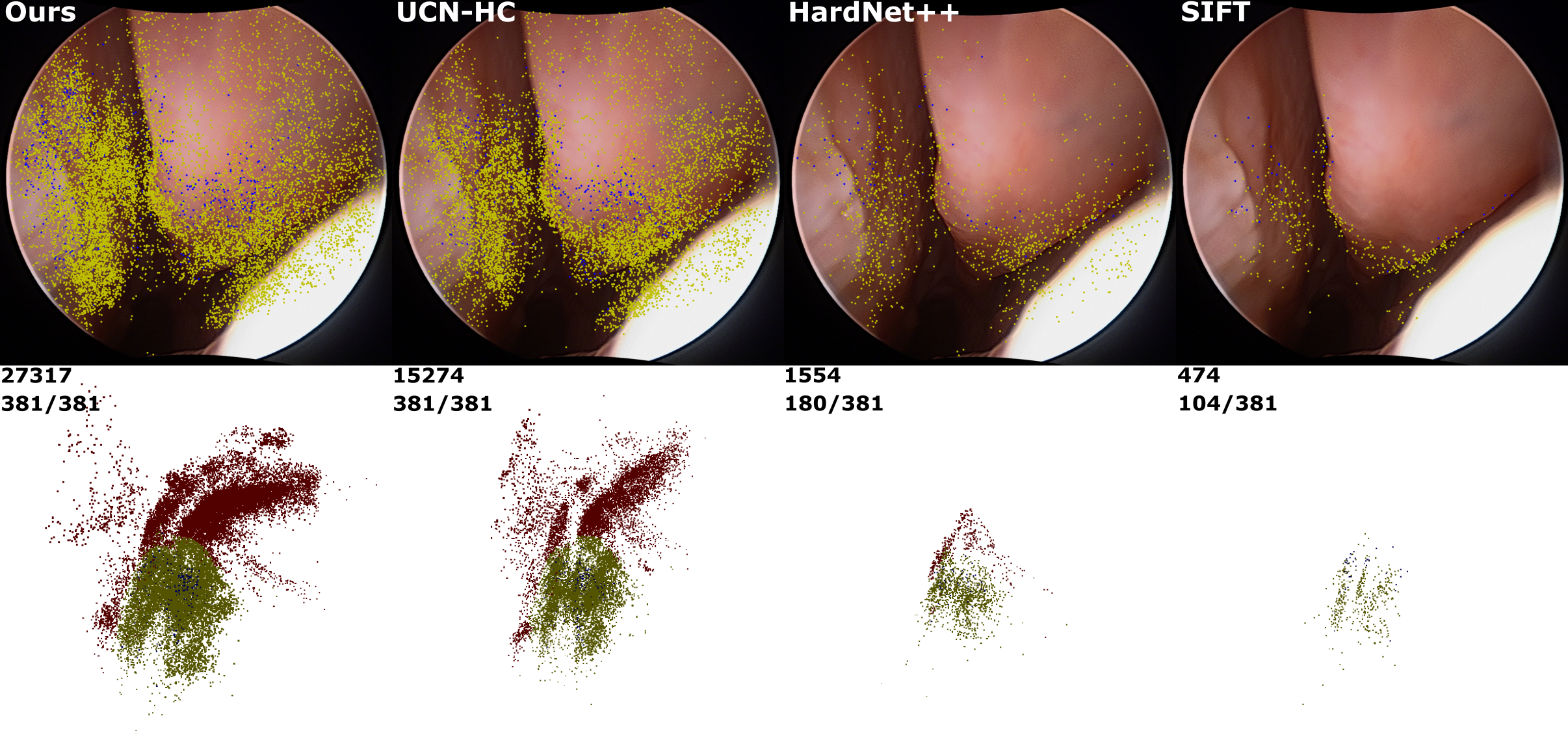
Extremely Dense Point Correspondences using Learned Feature
Descriptor
In this work, we present an effective self-supervised
training scheme and novel loss design for dense descriptor
learning. In direct comparison to recent local and dense
descriptors on an in-house sinus endoscopy dataset, we
demonstrate that our proposed dense descriptor can generalize
to unseen patients and scopes, thereby largely improving the
performance of Structure from Motion (SfM) in terms of model
density and completeness. We also evaluate our method on a
public dense optical flow dataset and a smallscale SfM public
dataset to further demonstrate the effectiveness and
generality of our method. (CVPR 2020)
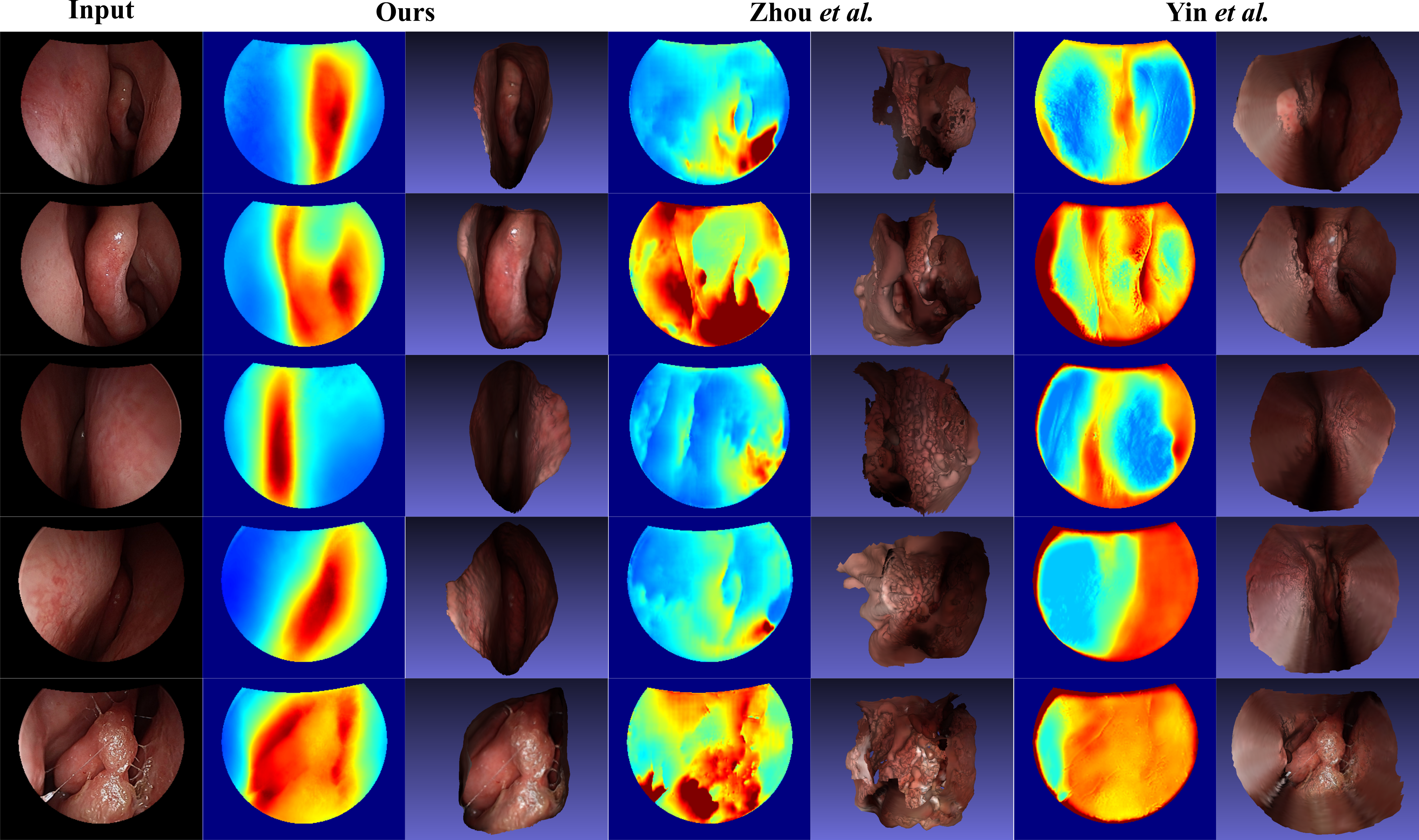
Dense Depth Estimation in Monocular Endoscopy with
Self-supervised Learning Methods.
We present a self-supervised approach to training
convolutional neural networks for dense depth estimation from
monocular endoscopy data without a priori modeling of anatomy
or shading. Our method only requires monocular endoscopic
videos and a multi-view stereo method, e. g., structure from
motion, to supervise learning in a sparse manner.
Consequently, our method requires neither manual labeling nor
patient computed tomography (CT) scan in the training and
application phases. In a cross-patient experiment using CT
scans as groundtruth, the proposed method achieved
submillimeter mean residual error. In a comparison study to
recent self-supervised depth estimation methods designed for
natural video on in vivo sinus endoscopy data, we demonstrate
that the proposed approach outperforms the previous methods by
a large margin. (IEEE TMI 2020)
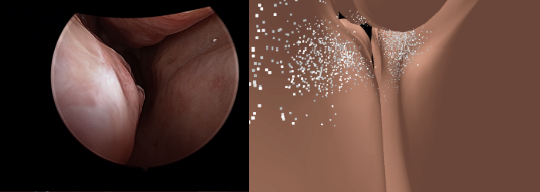
Endoscopic navigation in the absence of CT imaging
Clinical examinations that involve endoscopic exploration of
the nasal cavity and sinuses often do not have a reference
image to provide structural context to the clinician. In this
paper, we present a system for navigation during clinical
endoscopic exploration in the absence of computed tomography
(CT) scans by making use of shape statistics from past CT
scans. Using a deformable registration algorithm along with
dense reconstructions from video, we show that we are able to
achieve submillimeter registrations in in-vivo clinical data
and are able to assign confidence to these registrations using
confidence criteria established using simulated data. (MICCAI 2018)
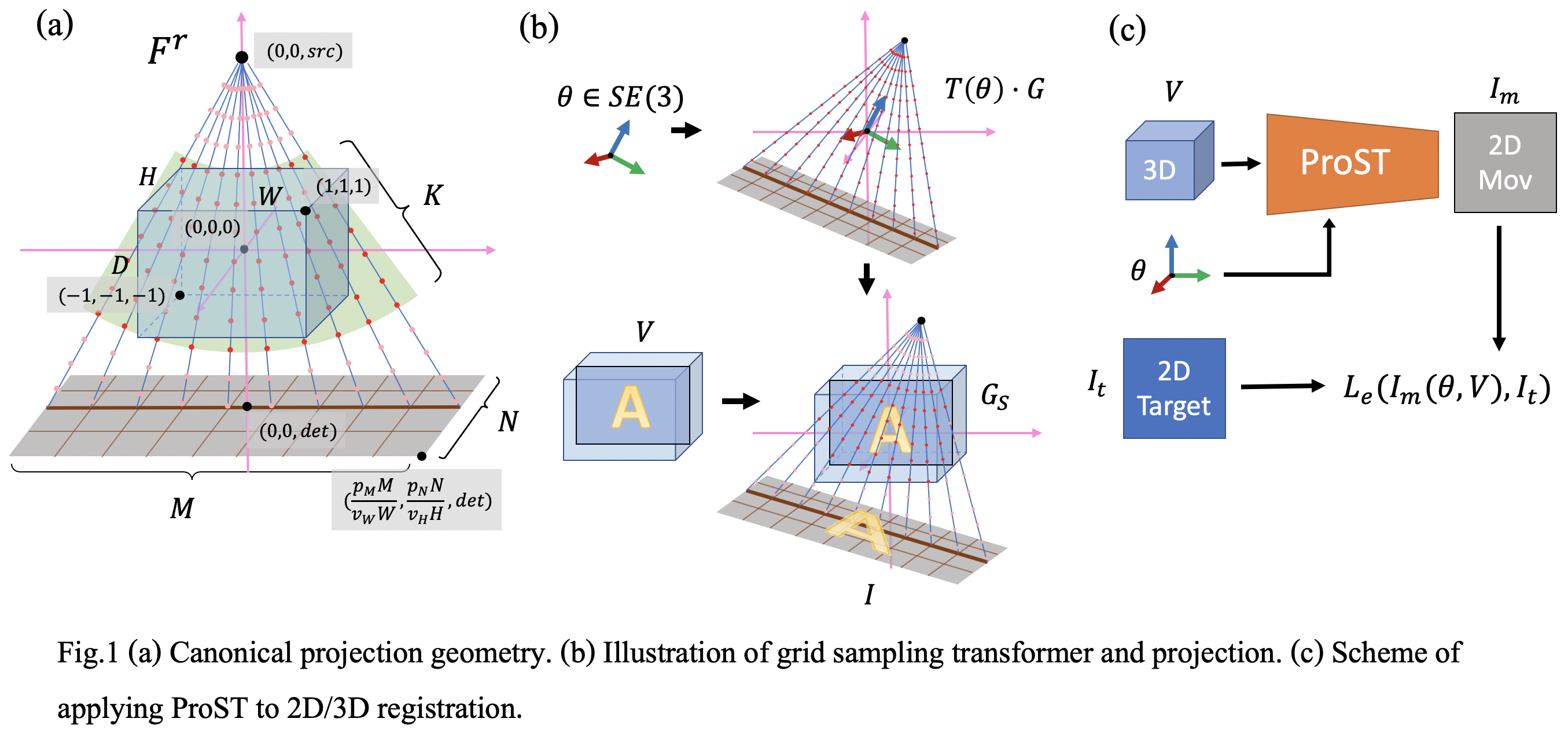
Generalizing Spatial Transformers to Projective Geometry
with Applications to 2D/3D Registration
Differentiable rendering is a technique to connect 3D scenes with corresponding 2D images. Since it is differentiable, processes during image formation can be learned. Previous approaches to differentiable rendering focus on mesh-based representations of 3D scenes, which is inappropriate for medical applications where volumetric, voxelized models are used to represent anatomy. We propose a novel Projective Spatial Transformer module that generalizes spatial transformers to projective geometry, thus enabling differentiable volume rendering. We demonstrate the usefulness of this architecture on the example of 2D/3D registration between radiographs and CT scans. Specifically, we show that our transformer enables end-to-end learning of an image processing and projection model that approximates an image similarity function that is convex with respect to the pose parameters, and can thus be optimized effectively using conventional gradient descent. To the best of our knowledge, this is the first time that spatial transformers have been described for projective geometry. The source code will be made public upon publication of this manuscript and we hope that our developments will benefit related 3D research applications. (MICCAI 2020)
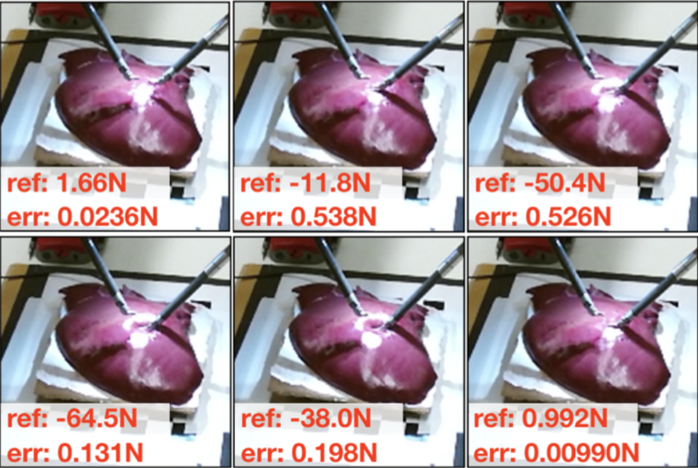
Learning to See Forces: Surgical Force Prediction with
RGB-Point Cloud Temporal Convolutional Networks
Robotic surgery has been proven to offer clear advantages
during surgical procedures, however, one of the major
limitations is obtaining haptic feedback. Since it is often
challenging to devise a hardware solution with accurate force
feedback, we propose the use of "visual cues" to infer forces
from tissue deformation. Endoscopic video is a passive sensor
that is freely available, in the sense that any
minimally-invasive procedure already utilizes it. To this end,
we employ deep learning to infer forces from video as an
attractive low-cost and accurate alternative to typically
complex and expensive hardware solutions. First, we
demonstrate our approach in a phantom setting using the da
Vinci Surgical System affixed with an OptoForce sensor.
Second, we then validate our method on an ex vivo liver organ.
Our method results in a mean absolute error of 0.814 N in the
ex vivo study, suggesting that it may be a promising
alternative to hardware based surgical force feedback in
endoscopic procedures. (MICCAI workshop 2018)

An Optical Tracker Based Registration Method Using Feedback
for Robot-Assisted Insertion Surgeries
Robot-assisted needle insertion technology is now widely used
in minimally invasive surgeries. The success of these
surgeries highly depends on the accuracy of the position and
orientation estimation of the needle tip. In this paper, we
proposed an optical tracker based registration method for the
marker- robot-needle tip registration in a robot-assisted
needle insertion surgery. The method consists of two steps.
First, with the guide of the optical tracker, we use the
motion vector from the marker’s current position to its target
point as a feedback control to automatically register the
robot-marker and the robot-tracker coordinates respectively.
Then, in the procedure of marker-needle tip registration, we
use two points on the needle holder to achieve a high accuracy
needle orientation registration. We have conducted a series of
experiments for the proposed method on a verified platform.
Results show that the position estimation error is below 0.5mm
and the orientation estimation error is below 0.5 degrees. (The
implementation and debugging of this work, e.g. GUI and
algorithms, are completed by myself alone. Zhuo Li is
responsible for evaluation experiment setup and paper
writing.)







