Linux Kernel 2.6.29 + tux3
In this section we are going to explore a certain version of the Linux Kernel, the tux3 branch from March 14, 2009. This version contains the Linux tree up to March 10. The 2.6.29 was released on March 23 so this is not exactly the final version. I picked it because it contains btrfs and, at the time of writing this report, it is the latest one that was published by Daniel Phillips, the creator of the tux3.
The rest of this section is made up exclusively of figures accompanied by extensive captions.

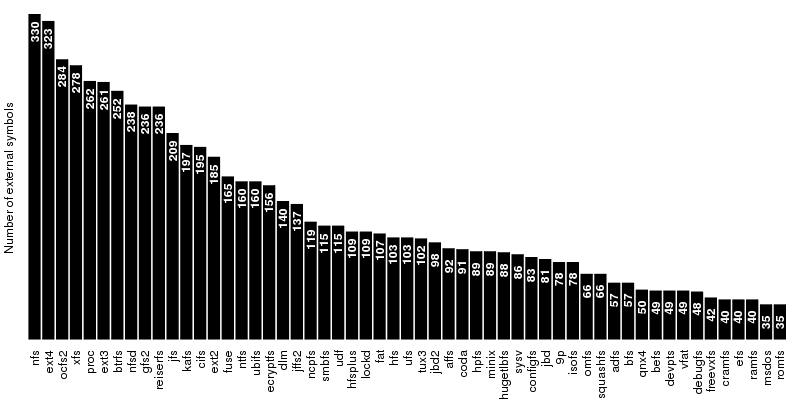
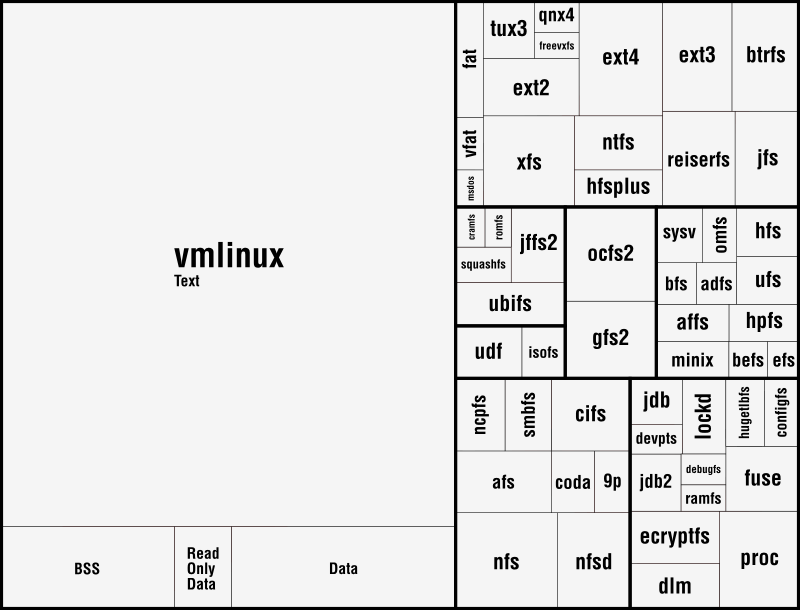
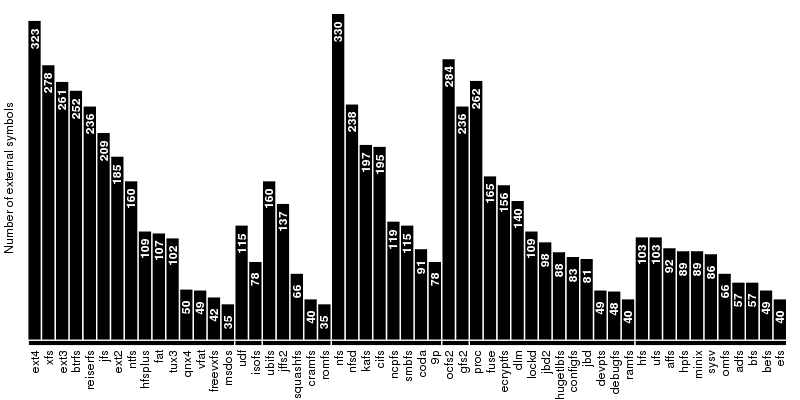
The first group, which contains the disk-based file systems, is led by ext4 which is ahead of xfs by 45 external symbols. Note that the number of external symbols for both ext4 and ext3 is boosted by the fact the journaling part not implemented internally but provided by jbd2 and jbd respectively. At the other end of the scale is a group of 4 file systems out of which only two, freevxfs and qnx4 are truly self-contained. The other two, msdos and vfat are getting most of their functionality from the fat module which is more than twice of their size.
The second group contains the file systems dedicated to optical mediums and it holds no surprises: udf is ahead of isofs by a comfortable margin.
The same thing is also true for the the third group, of the flash-based file systems, where the first place is taken by ubifs followed by jffs2. The third placed is secured by squashfs while the bottom is shared by cramfs and romfs which are separated by only 5 symbols.
The fourth group is the one dedicated to the network file systems. Here the first two spots are taken by nfs and nfsd, which provide kernel support for NFS client and NFS server. On the next two places, at very close distance between them, are kafs, the Andrew File System, and cifs. The end is shared by coda and 9p.
The fifth group contains, in this order, the only two cluster-based file systems: ocfs2 and gfs2. The number of external symbols for ocfs2 is increased due to the use of the jbd2 journaling library.
The sixth group, the one dedicated to memory-based file systems is dominated authoritatively by proc which has almost 100 more external symbols than fuse, the file system from the second place. As expected, at the bottom sits ramfs.
The seventh and last group is dedicated to ancient file systems. The first place is shared by hfs and ufs. Quite surprising, this is only one of the three ties in this group, the other two being hpfs/minix and adfs/bfs.
The first two symbols are used by the function tracer. On the third place we have a tie between kfree, which is used by everybody except by ramfs, msdos and romfs, and kprintf which, surprising, is avoided by vfat, ramfs and msdos.
As expected, the kmem operations are among the most popular one. So are the some basic operations like strlen, memcmp and strsep.
Another thing we can notice are some (expected) pairs of calls that are always used together: register_filesystem/unregister_filesystem, _spin_lock/_spin_unlock and kmap/kunmap.
Another (again expected) observation is that the lack of (un)register_filesystem identifiers in the modules which only provides services to others: dlm, lockd, fat, and jbd2/jbd2.

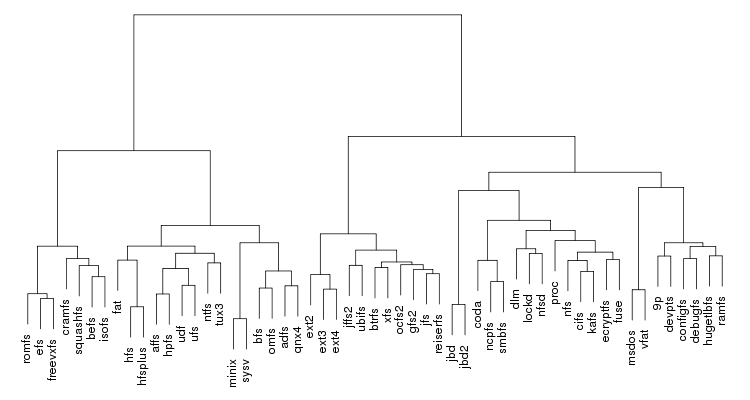
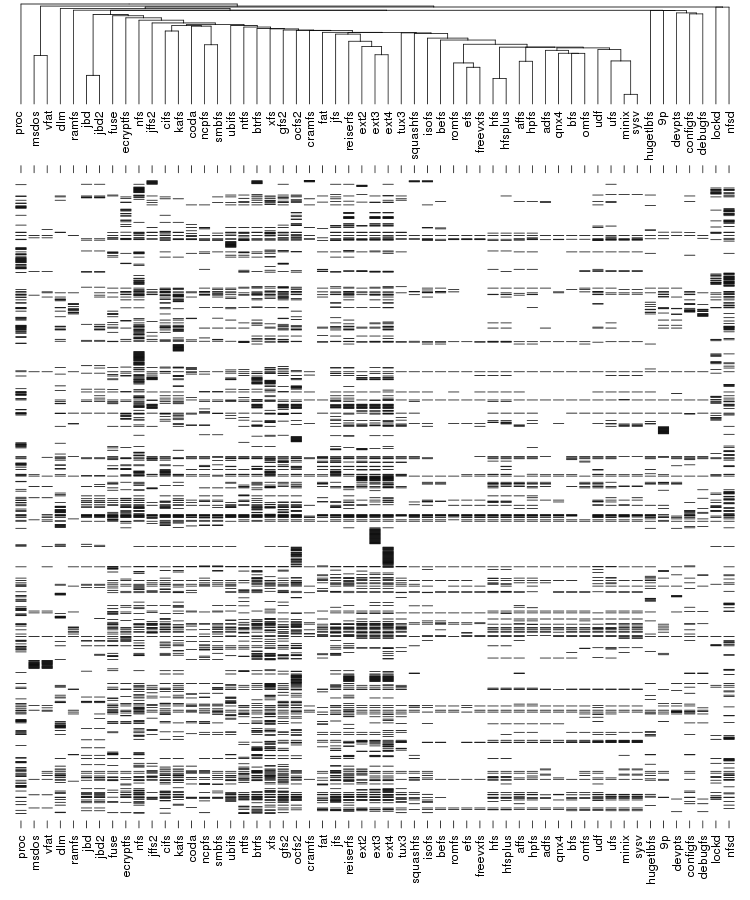
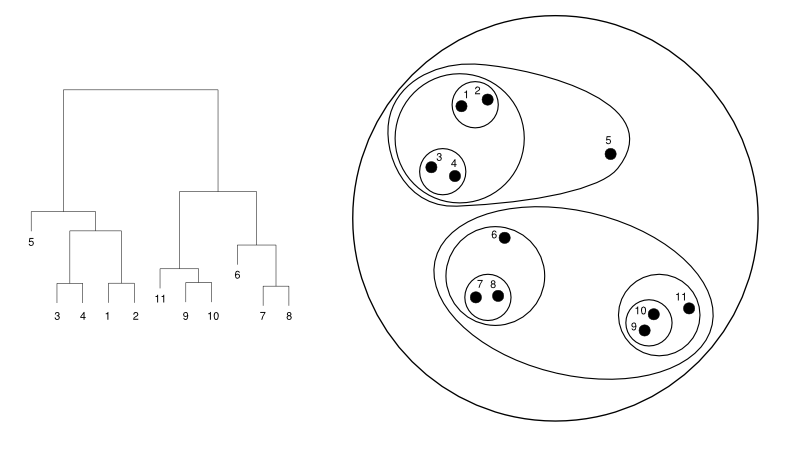
Below is an example of clustering 11 points situated in a 2D plane. Left is a dendrogram, a tree diagram usually used to represent the result of a hierarchical clustering. Right is a representation using nested clusters.
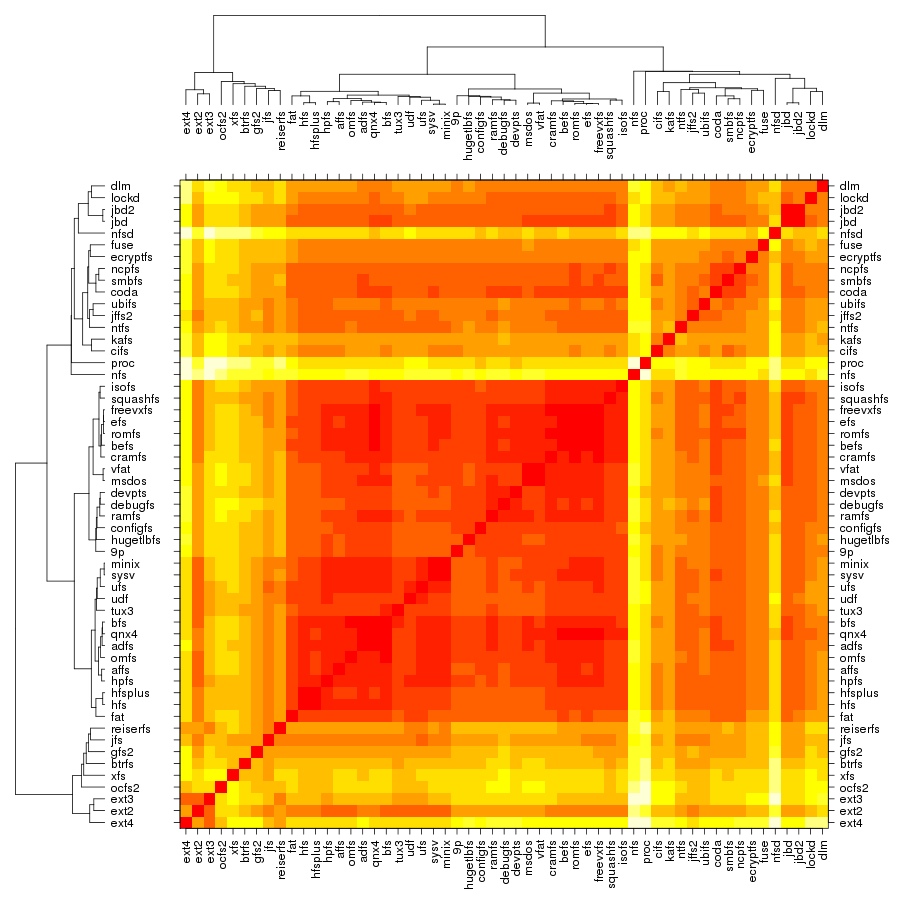
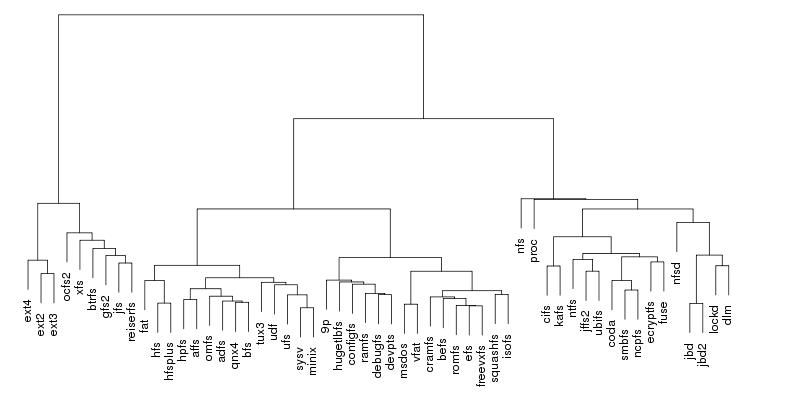
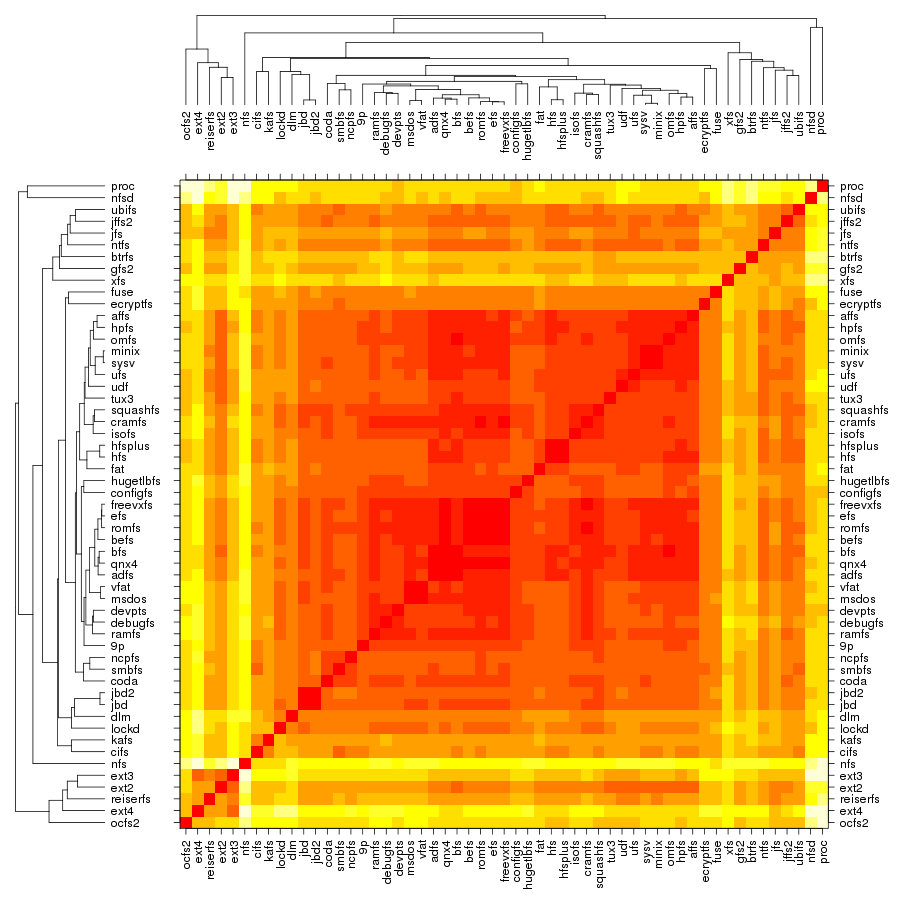
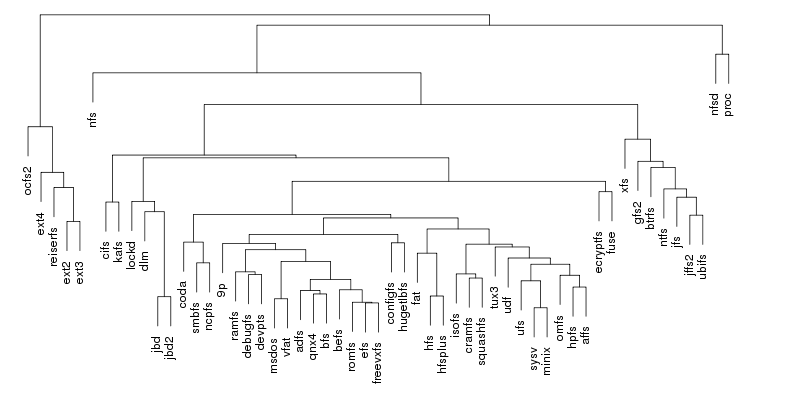
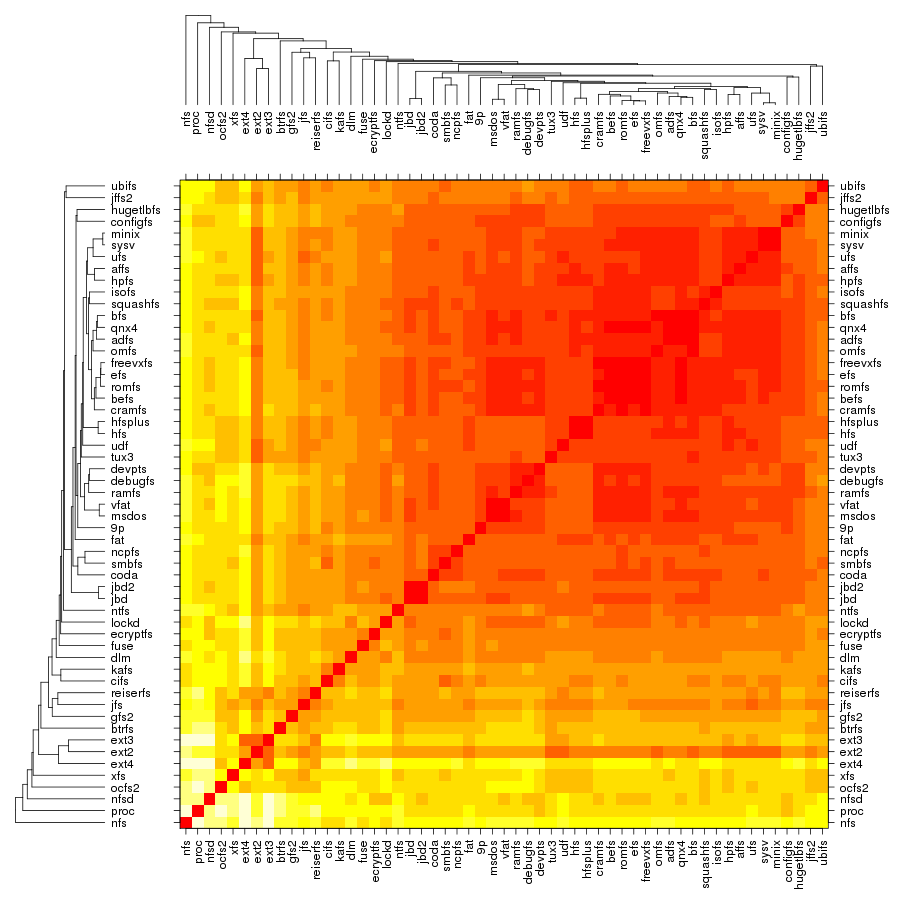
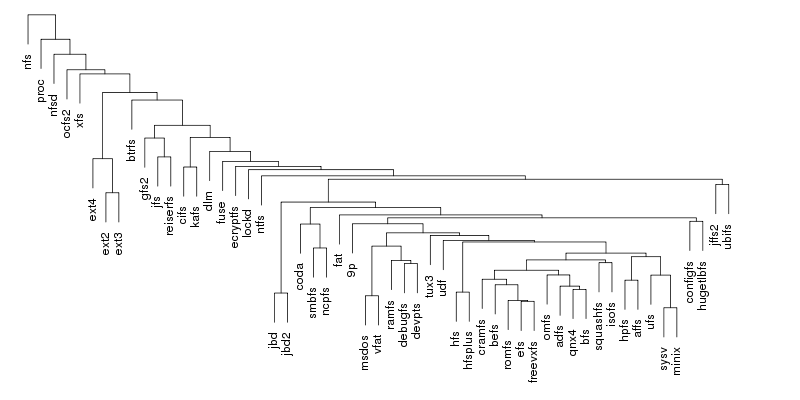
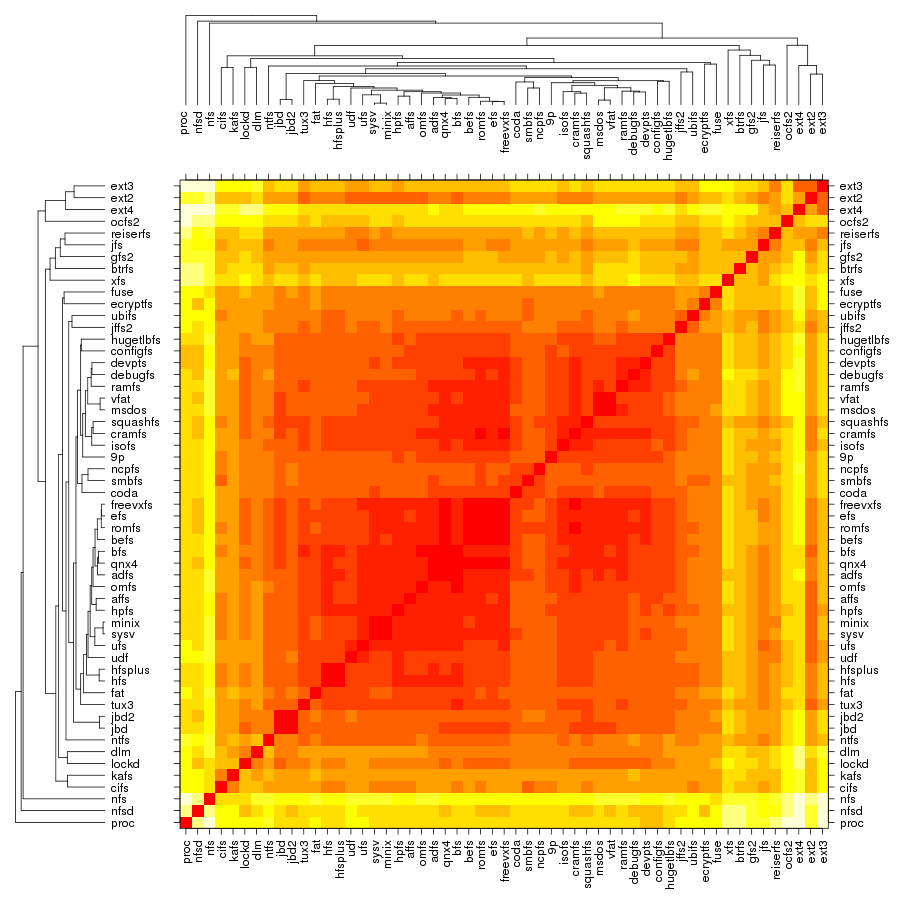
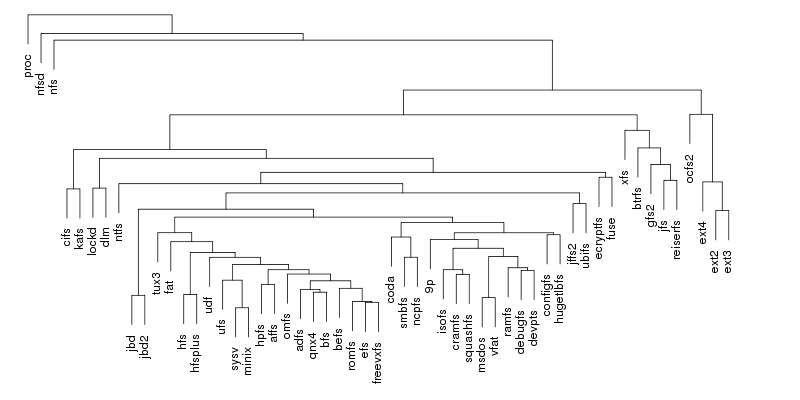
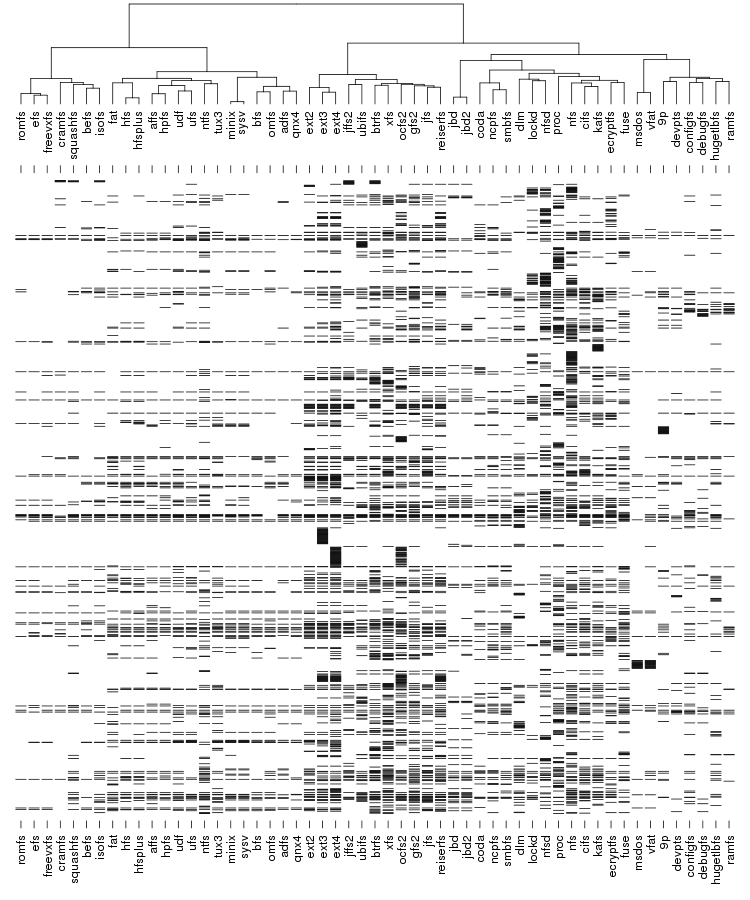
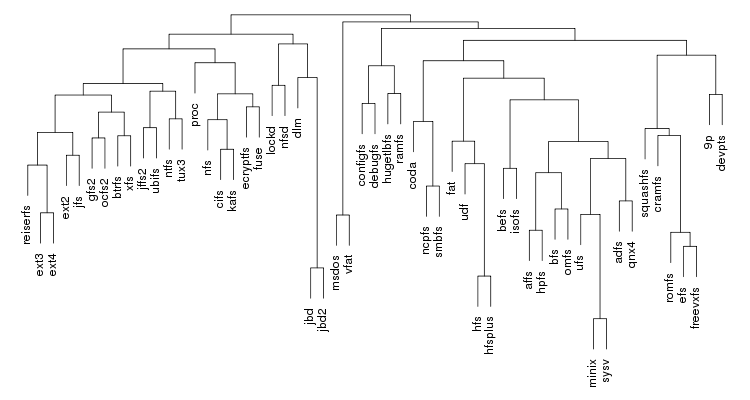
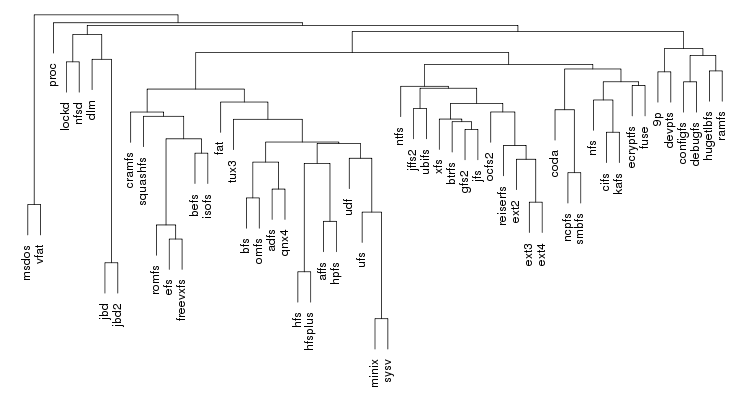
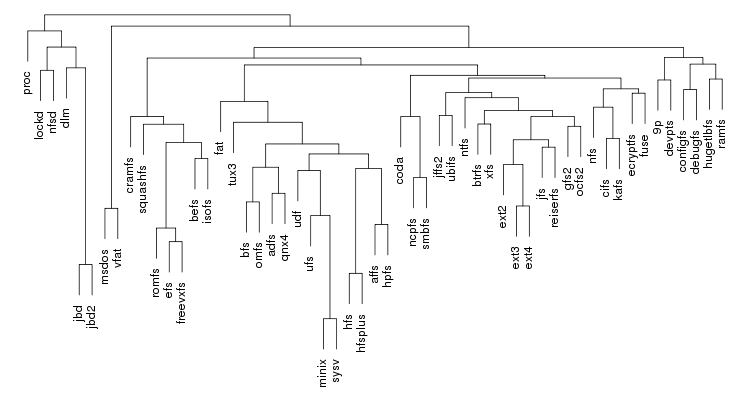
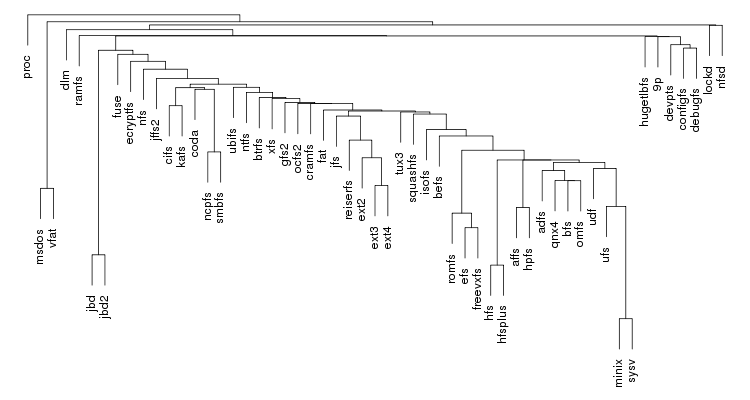
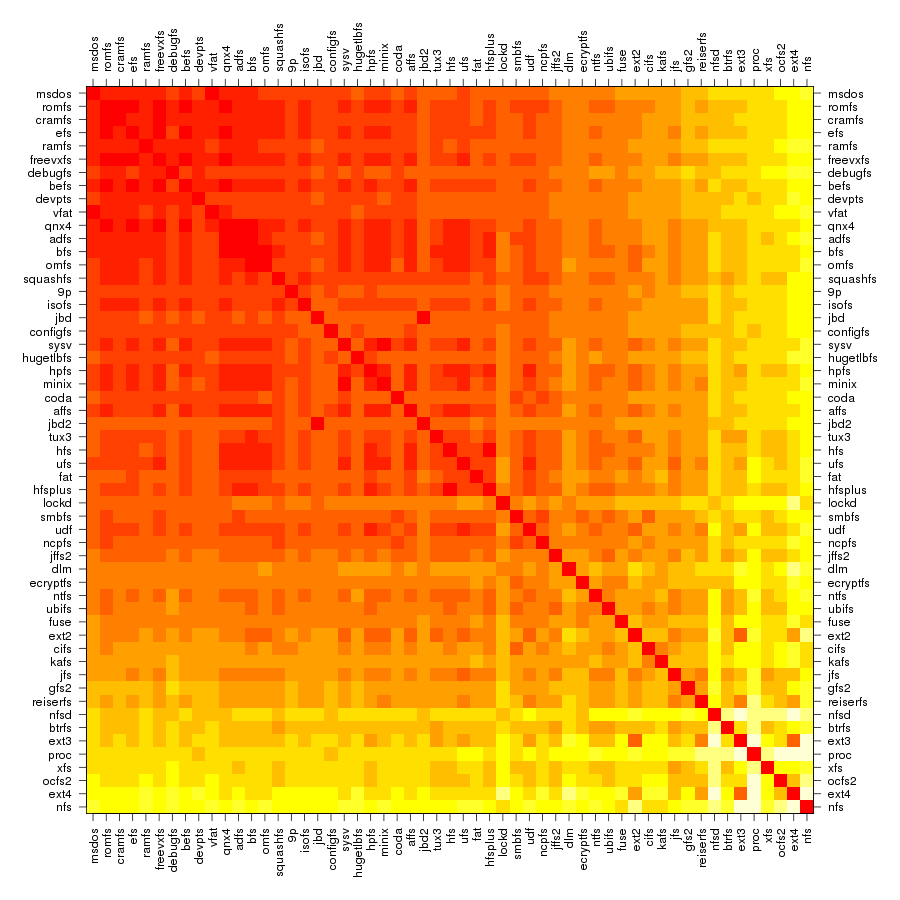
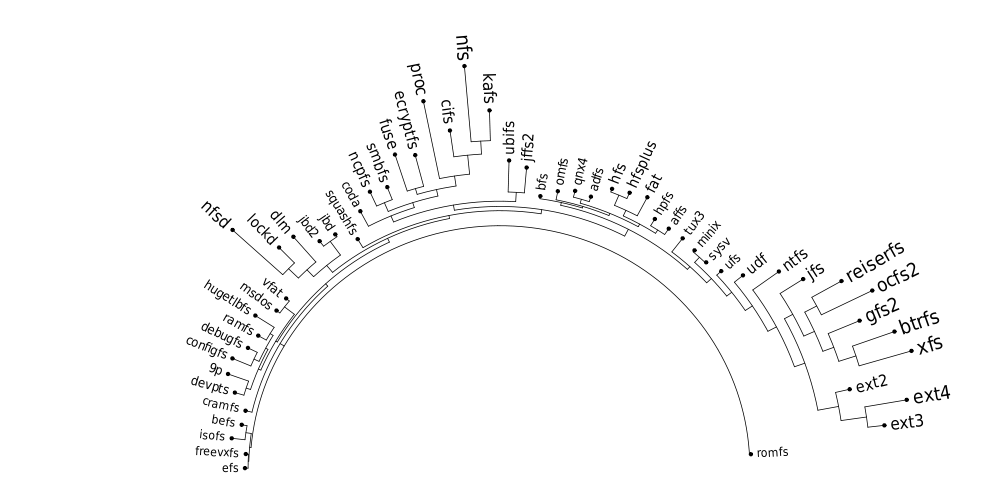
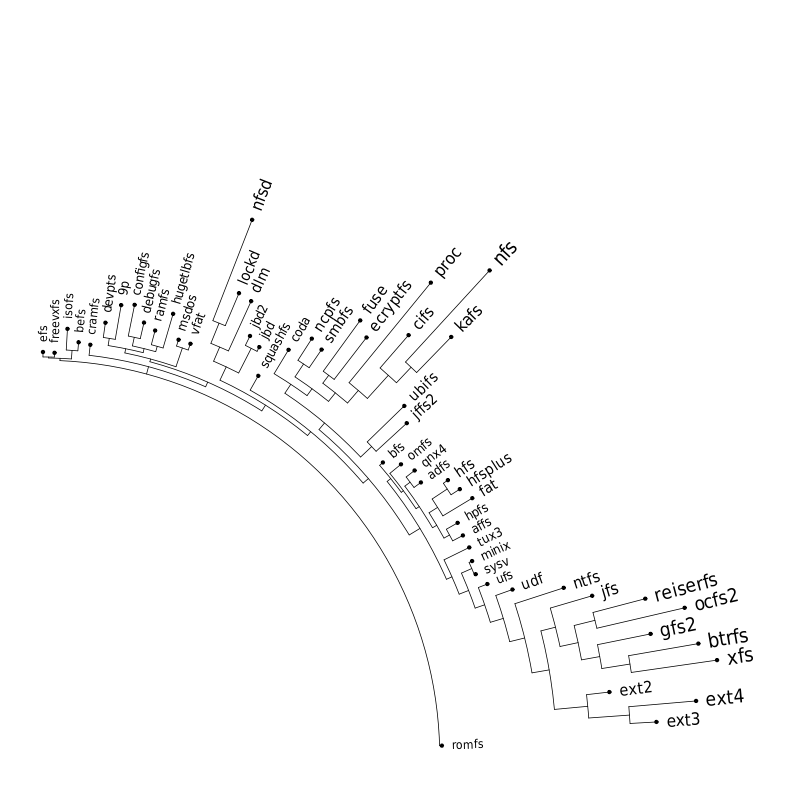
Following are a set of dendrograms using Canberra distance. In our case, this metric is equivalent with the number of different external symbols between two modules. After each dendrogram a reordered map of symbols is also plotted.
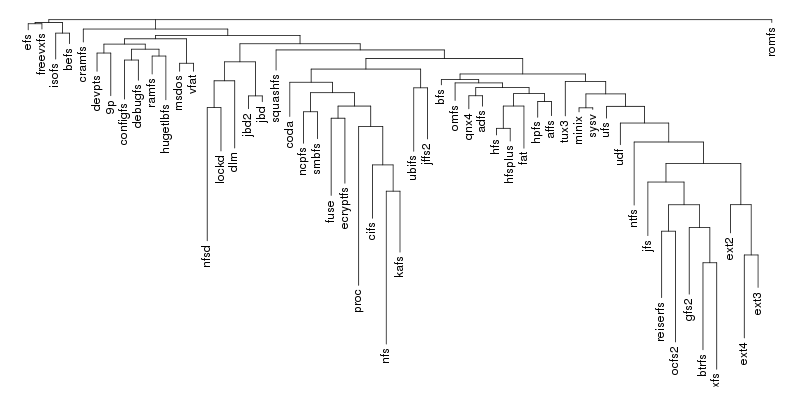
The input to Pars is number of species, each described using a string of characters. Usually each character is either "0" or "1" indicating the presence of absences of a certain feature but Pars also capable of dealing with up to 8 states plus "?" which indicates an unknown state. In our case we only need "0" and "1" and each position in the string encodes a certain external call.
The result is the following tree. This looks like a dendrogram but it is slightly different. This time the length of any vertical line is proportional with the number of changes between two states. We can see for example that msdos and vfat are both very close to the their parent while ext4 and ext3 are much farther apart.
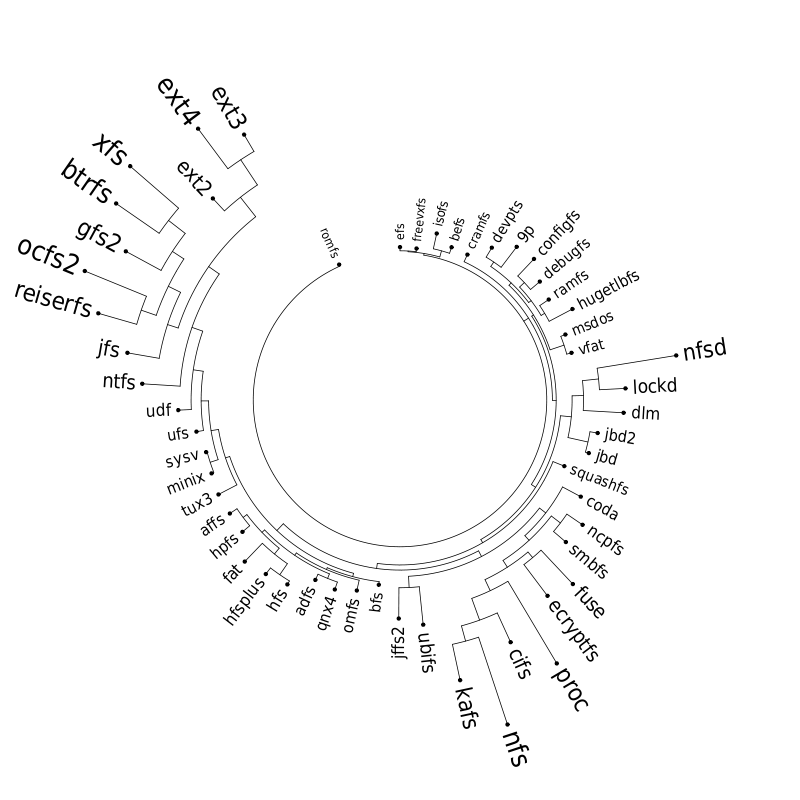
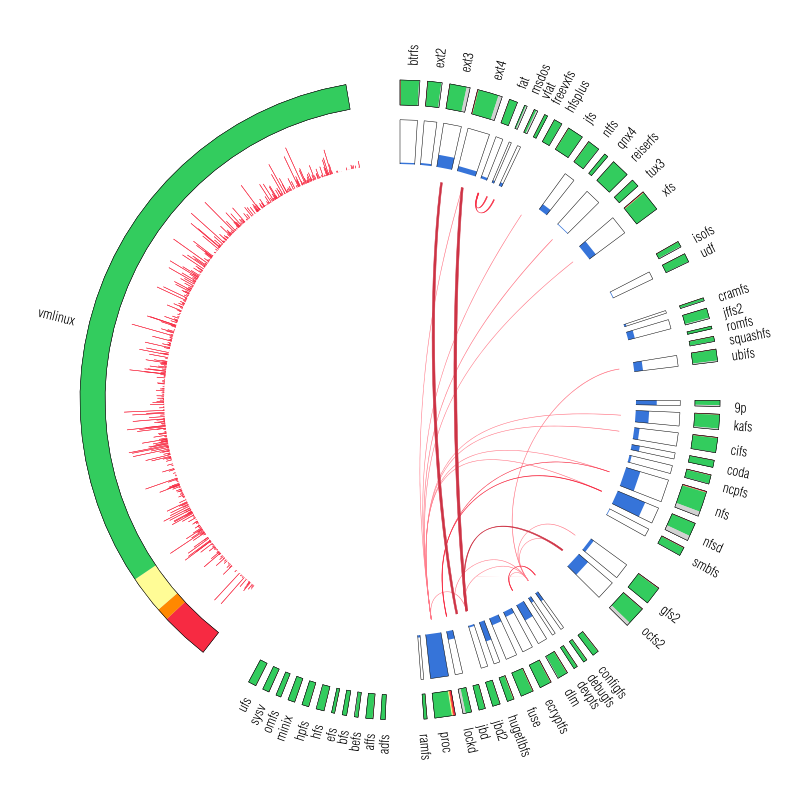
Let's now look at our plot. On the outer edge we have the file systems split in categories (from top to bottom: disk-based, optical mediums, flash-based, network-based, cluster-based, memory-based, ancient). The size is proportional with the number of external symbols. Colors indicate the type of external symbols. The green represents functions, red is data, orange are read-only data, and light yellow is uninitialized data (BSS). To give a sense of proportions, the external symbols exported by the Linux Kernel, vmlinux, are also depicted. It was compiled in the same configuration as the rest of the file systems. To give some numbers: it exports a total of 9310 external symbols out of which 8047 are functions, 621 are writable variables, 159 are read-only and 483 are BSS data. The gray area from file systems indicates the external symbols which are not satisfied by the kernel but by some other kernel module. This is noticeable for nfs, nfsd and also the users of jbd/jbd2: ext3, ext4, ocfs2.
On the inner edge of vmlinux there is a plot that indicates the frequency which which each exported symbol is used by the file systems from right. One thing we noticed here is that variables are used pretty much with the same frequency as the functions.
The set of boxes from the inner edge of the file systems represents the percentage of the external symbols which are unique to each file system. We can see that virtually all the external symbols used by proc are only used by it. But having unique external symbols is not a rare feature: with the exception of ancient file systems all the other categories have members with various degree of "uniqueness".
The red arcs from inside depict the use-provide relationships between the file systems. As expected, the memory-based modules are the ones that are the main providers with proc and debufgs being the most popular one. We can also see that lockd is used by nfs and nfsd and also the relation between fat and vfat/msdos. A notable thing: there is no link between dlm and ocfs2/gfs2 because only the main kernel module was considered.